Synthesis & Formulation
Drug delivery begins with the selection of a drug and complementary vehicle. In our lab we explored a large variety of drug carriers (home-made or through collaborations), however we are particularly focusing onto the world of self-assembly for nanomedicine. We work mainly on two carrier families:
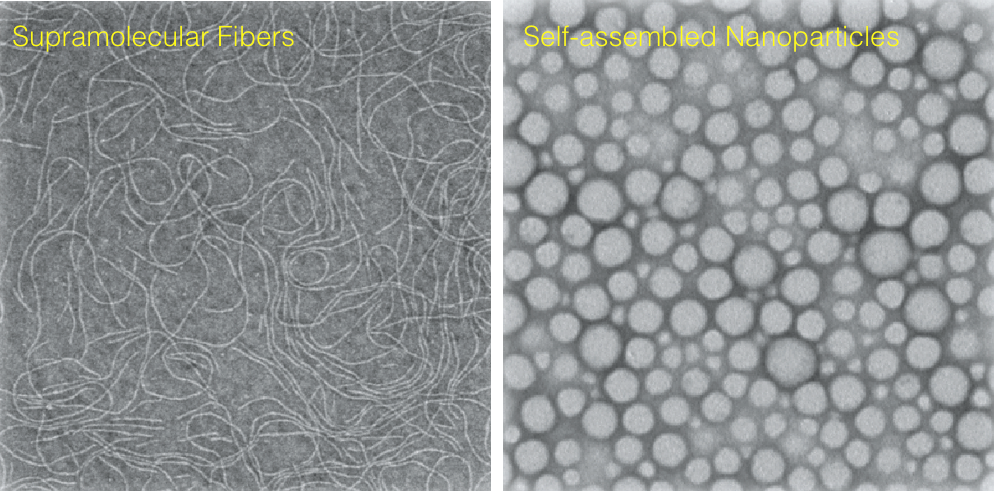
- synthesis of novel supramolecular assemblies (fibers),
- formulation of tunable PLGA-based drug nanocarriers (nanoparticles).
Supramolecular assemblies have been chosen for their versatility and modularity allowing the formulation of a large variety of carriers exploring a wide chemical-space of properties (size, shape, surface chemistry, degradability, functionality). Furthermore, supramolecular structures exhibit dynamic behavior: the non-covalent nature makes them ideal to respond to one or more trigger, changing the assembly state or promoting the disassembly, to expose or release a drug.
In our group we focus on the design of formulations that allows in a rapid and straight-forward manner to obtain a large library of materials. This is possible combining solid-phase monomer synthesis, automation and microfluidic devices for formulation.

Casellas, N.M., Pujals, S., Bochicchio, D., Pavan, G.M., Torres, T., Albertazzi, L., García-Iglesias, M., 2018. From isodesmic to highly cooperative: reverting the supramolecular polymerization mechanism in water by fine monomer design. Chem. Commun. 54, 4112–4115.
Characterization
To understand the properties and characteristics of the materials we synthesize and formulate we analyze them using different techniques. Most frequently we rely on a combination of standard techniques (HPLC, TEM, CD, DLS, spectroscopy) with in-house developed super-resolution imaging methods.
Super-resolution microscopy allows to visualize nanoparticles with single molecule sensitivity and therefore allows to study materials and their composition and properties with unprecedented resolution. Combining smart labeling with techniques such STORM and PAINT it is possible to measure relevant properties (size, shape, molecular composition and functionality) at the single nanoparticle level and therefore study structural and functional heterogeneity.
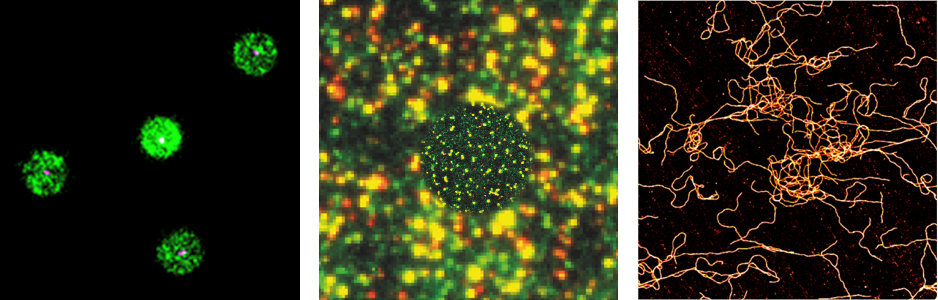

Albertazzi, L., Zwaag, D. van der, Leenders, C.M.A., Fitzner, R., Hofstad, R.W. van der, Meijer, E.W., 2014. Probing Exchange Pathways in One-Dimensional Aggregates with Super-Resolution Microscopy. Science 344, 491–495. https://doi.org/10.1126/science.1250945
Feiner-Gracia, N., Olea, R.A., Fitzner, R., El Boujnouni, N., van Asbeck, A.H., Brock, R., Albertazzi, L., 2019. Super-resolution Imaging of Structure, Molecular Composition, and Stability of Single Oligonucleotide Polyplexes. Nano Lett. 19, 2784–2792.
Circulation, Extravasation & Diffusion
We use advanced optical microscopy techniques (super-resolution imaging, single molecule tracking, spectral imaging) to track the nanomaterials in their journey in the body understanding their behavior and their interactions in the biological environment. This is key to rationally design improved carriers with controlled behavior.
Circulation
The majority of nanocarriers under development are designed to be injected intravenously. This delivery route has an advantage of lowering the drug dose and faster action, however it implicates a number of biological interactions with serum proteins, lipids and small metabolites, as well as a variety of circulating cells. The understanding and control of such interactions is key as they can result in the inactivation of the drug carriers.
Here we use advanced optical imaging techniques to understand key phenomena occurring during circulation such as protein corona formation, carriers’ stability and degradation and immune response. The heterogenous and complex nature of these phenomena and the small scale of nanoparticles make these studies challenging. We introduce the use of super-resolution techniques such as STORM to complement the existing methods allowing single-particle nanoscale analysis of nano-bio interactions.
Extravasation
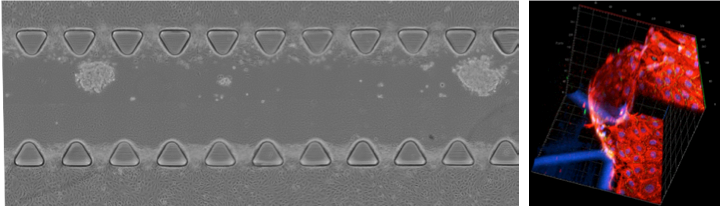
The selective extravasation of nanocarriers from the vasculature to the cancer tissue is a crucial step to achieve enhanced tumour accumulation. Therefore, NPs must reach the vessel wall and be able to cross the endothelial barrier.
Most of the nanocarriers under development rely on the Enhanced-Permeation Retention (EPR) effect to effectively escape the circulation in a blood vessel and reach the tumour malignant cells. However, it has been recently reported that the EPR effect is oversimplified and heterogenous among the patients.
One of the major limitations of current investigations is the fact that 2D in vitro models do not provide exhaustive information, as they do not fully recapitulate the 3D tumor microenvironment.
We work on real-time monitoring of extravasation in a 3D environment by combining live cell imaging with organ-on-a-chip microfluidic models. This allows to understand the behavior of particles at the endothelial barrier and to screen different formulations with tuned properties to promote selective extravasation.
Diffusion
After leaving the bloodstream, nanomaterials encounter a very different microenvironment. Indeed, the extracellular matrix (ECM) is a complex 3D network of biological macromolecules that regulates the mobility of cells and molecule within the intracellular space. The ECM properties (mesh size, charge) depends on the tissue and can severely affect the ability of nanoparticles to reach the cancer cells. In our group we are performing diffusion studies of nanoparticles within tunable models of extracellular matrix using a variety of biophysical techniques (spectral imaging, FRAP, single particles tracking). This information will contribute to draw a map of structure-diffusion relationship and to design carriers able to reach more easily the target site.

Feiner-Gracia, N., Beck, M., Pujals, S., Tosi, S., Mandal, T., Buske, C., Linden, M., Albertazzi, L., 2017b. Super-Resolution Microscopy Unveils Dynamic Heterogeneities in Nanoparticle Protein Corona. Small 13. https://doi.org/10.1002/smll.201701631
Feiner-Gracia, N., Buzhor, M., Fuentes, E., Pujals, S., Amir, R.J., Albertazzi, L., 2017a. Micellar Stability in Biological Media Dictates Internalization in Living Cells. J. Am. Chem. Soc. 139, 16677–16687. https://doi.org/10.1021/jacs.7b08351
Feiner-Gracia, N., Glinkowska Mares, A., Buzhor, M., Rodriguez-Trujillo, R., Samitier, J., Amir, R.J., Pujals, S., Albertazzi, L., 2020. Real-time Ratiometric Imaging of Micelles Assembly State in a Microfluidic Cancer-on-a-chip. bioRxchiv
Cellular Targeting
Active targeting consists of functionalizing nanocarriers with ligands able to specifically bind to biomarkers present on the disease cell surface. This strategy has been projected as the magic bullet to effectively differentiate cancer cells from healthy ones and, consequently, selectively direct a nanocarrier to its site of action.
Despite the excitement about active targeting there is no approved active targeted nanocarrier in the clinic up to date. One of the reasons is that the targeting phenomenon is often oversimplified while reality is more complex. For example the overexpression of markers by cancer cells is not a black or white condition, and healthy cells also present the targeted protein. Moreover, the high variability between patients and within tumours enhance the difficultly to optimize these carriers.
Looking at the design of a targeted carrier many parameters have to be taken in account: size, nature and number of ligands and of course their affinity. Therefore the rational design of specific nanoparticles is far from trivial.
In our group we tackle this issue from multiple points of view. First we use nanoscopy to look at the number and nanoscale distribution of cancer cells biomarkers with high sensitivity and resolution. Second we use super-resolution imaging to obtain functional information of targeting nanoaprticles (see also characterization section). Finally, we are active in the synthesis of targeting ligands (e.g. peptides) with tailored affinity and target.
Combining the knowledge of cell receptor imaging (see section Receptor imaging) and the characterization of targeting ligands on nanomaterials we intent to design personalized nanocarriers with improved targeting efficiency.
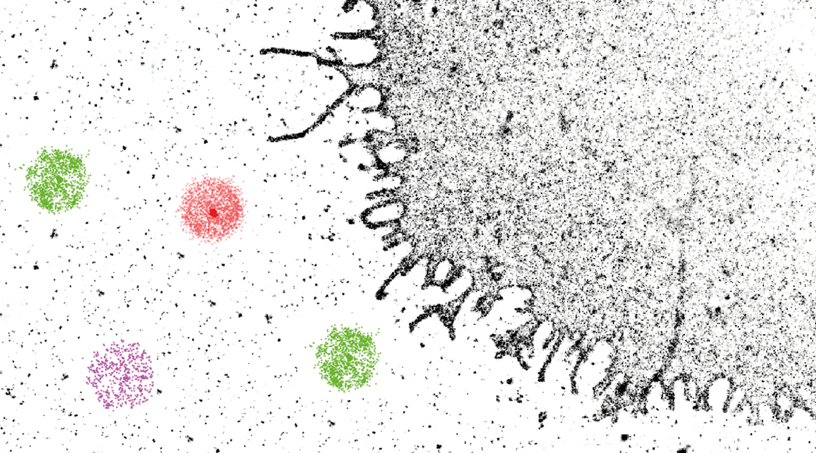

Delcanale, P., Miret-Ontiveros, B., Arista-Romero, M., Pujals, S., Albertazzi, L., 2018. Nanoscale Mapping Functional Sites on Nanoparticles by Points Accumulation for Imaging in Nanoscale Topography (PAINT). ACS Nano 12, 7629–7637.
Intracellular Localization & Activity
Understanding nanoparticles cell localization and trafficking after membrane binding is of crucial importance. To address these issues, we use an array of nanoscopy methods to track nanoparticles inside the cell. Using multicolor nanoscopy we can accurately localize nanoparticles position and co-localization with intracellular structures. This is key to identify trafficking pathways. Moreover, with a smart labeling we can assess carrier functionalities such as drug release and carrier stability. This is done with a combination of high-resolution static techniques (STORM, PAINT) with high-temporal resolution single molecule imaging (single-molecule tracking). Moreover, correlative microscopy (CLEM) is used to visualize the cell ultrastructure (by electron microscopy) and the localization of the molecule of interest (by optical nanoscopy).
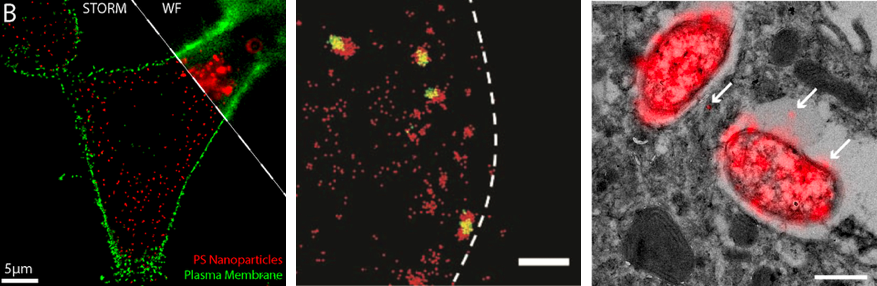

van Elsland, D.M., Pujals, S., Bakkum, T., Bos, E., Oikonomeas‐Koppasis, N., Berlin, I., Neefjes, J., Meijer, A.H., Koster, A.J., Albertazzi, L., van Kasteren, S.I., 2018. Ultrastructural Imaging of Salmonella–Host Interactions Using Super‐resolution Correlative Light‐Electron Microscopy of Bioorthogonal Pathogens. Chembiochem 19, 1766–1770. https://doi.org/10.1002/cbic.201800230
Riera, R., Feiner-Gracia, N., Fornaguera, C., Cascante, A., Borrós, S., Albertazzi, L., 2019. Tracking the DNA complexation state of pBAE polyplexes in cells with super resolution microscopy. Nanoscale 11(38):17869-17877.